Introduction
Environmental pollution of heavy metals is increasingly becoming a problem and has become of great concern due to the adverse effects it is causing around the world. Because of the rapidly developing agriculture and metal industries, these inorganic pollutants are being wasted in our waters, soils, and environment (Wolińska et al., 2013). Some metals interfere with bio-rescales functioning and growth, while other wises building on in one or more organs, resulting in a variety of dangerous disorders such as cancer, delayed nerve responses, mutagenic changes and neurological issues (Briffa et al., 2020).
Chromium (Cr) is a naturally occurring element found in the earth’s crust that has oxidation states ranging from chromium (III) to hexavalent chromium (Monalisa and Kumar, 2013). Chromium is released into multiple environmental matrices (air, water, and soil) through a range of natural and anthropogenic sources. Originating in industrial settings, metal processing, tannery facilities, chromate manufacturing, stainless steel welding, and ferrochrome and chrome pigment production are the industries that contribute the most to chromium release (Jacobs and Testa, 2005). The health risk associated with chromium exposure is determined by its oxidation state, which ranges from minimal toxicity in the metal form to severe toxicity in the hexavalent form. Humans, plants, animals, and fish are all at risk from hexavalent chromium compounds. Chromium has been categorized as a Priority pollutant or Class A pollutant by the United States Environmental Protection Agency (USEPA) due to its carcinogenicity and mutagenicity (Srinath et al., 2002). Heavy metals, such as chromium, harm cell membranes, change enzyme specificity, disrupt cellular activities, and destroy deoxyribo nucleic acid (DNA) structure at high doses (Ghani, 2011).
Chromium is a recognized mutagen, producing mitotic obstruction, cell growth inhibition, and cell death. It can cause oxidative damage to DNA by generating free radicals if it is not reduced immediately. Hexavalent chromium has been shown to induce lung cancer, chromate ulcers, nasal septum apertures, and kidney damage in humans, as well as harm to other living things (Bruins et al., 2000).
The hexavalent chromium can be reduced Cr (III) from the environment through reduction or absorption techniques. These procedures are costly, energy-intensive and they run the danger of secondary chemical contamination. Heavy metal-contaminated wastewater and effluents can be treated using possible microorganisms as an alternative technique. These microorganisms can treat huge quantities of wastewater for a cheap operating cost and low energy consumption, as well as better metal removal efficiency (Zayed and Terry, 2003).
In the region where industrial effluents are discharged, several microorganisms can defend themselves against the toxicity of heavy metals present in the effluents. These microorganisms, which include bacteria, fungi, algae, and protozoa, employ a variety of strategies to survive heavy metal toxicity, including heavy metal absorption, adsorption oxidation, methylation, and heavy metal reduction to benign forms. Many species employ hazardous Cr (III) reduction to nontoxic Cr (III) as one of their survival strategies in Cr (III) polluted effluents. Heavy metal bioremediation by bacterial cells has been identified as a viable alternative to conventional heavy metal technologies from industrial wastes (Ahluwalia and Goyal, 2007). This is an attempt to explore innovative, cost effective and environment friendly technology for the bioremediation of Cr (VI) contamination using microorganisms. Therefore, the present study aims to isolate of different isolates of Pseudomonas spp. from different industrial wastewater sources contaminated with chromium and evaluate the efficiency of the chromium tolerant Pseudomonas species in bio remediating chromium contaminated industrial wastewater.
Materials and Methods
Stock solution of Cr (VI) was prepared by dissolving K2Cr2O7 (Lanxess, Köln, Germany) in microbiological grade water. A certain volume of Cr (VI) from the stock solution was taken, sterilized and added into the media before inoculation to obtain the desired Cr (VI) concentration; all medium components, di-phenyl carbazide substance (DPC) and chemicals used in this study were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Ten industrial waste water samples from different leather tanneries area in Fayoum governorate, Egypt were collected and the physicochemical parameters of wastewater viz., temperature (°C), pH, dissolved oxygen and chromium (μg/mL) were measured according to the method of APHA (1989) and stored at 4°C until the analysis was carried out. For isolating chromium tolerant bacteria, one mL of water samples were first mixed with nine mL of distilled water in a test tube and heated for 15 minutes at 65°C in a hot water bath. Serial dilutions from 10–1 to 10–10 were prepared using sterilized saline solution. An aliquot of 100 μL of each dilution was spread on king B agar plates, pH 7.0 and incubated at 37°C for 24–48 h (Ben-David and Davidson, 2014). The isolated single cells were preserved in 40% glycerol solution and kept at –80°C for further identification and screening for Cr tolerant bacterial isolates.
Bacterial strains were tested for their resistance to chromium. The selected isolates related Pseudomonas spp after morphological and biochemical tests were grown in 50 culture Luria- Bertani (LB) medium containing the following components (g/L): Tryptone (10.0), NaCl (10.0) and yeast extract (5.0) at pH 7. 100 μL/mL of chromium concentration was added to the medium and incubated at 37°C at 3.77 ×g for 24 h. The selected cultures displaying the highest bacterial growth were selected for further characterization and were maintained on nutrient agar slants and stored at 4°C.
The effect of bioremediation bacterial growth was determined by bacterial growth curve using spectrophotometer at 600 nm according to Poornima et al. (2010). For estimation of bacterial isolates activity, the isolates were grown in LB medium containing different concentrations of chromium (100, 200, 300, 400, 500, 600, 700, 800, 900, 1,000, and 1,200 μg/mL).
The bactericidal activity of Cr (VI) was studied by determining the minimum inhibitory concentration (MIC) by the broth dilution method (Calomiris et al., 1984).
Chromium (VI) reduction potential of this strains were investigated by1,5-diphenylcarbazide (DPC) method (APHA, 1989), in this method, Cr (VI) was reacted with 1,5-diphenylcarbazide (DPC) dye, which, in acidic conditions, forms a purple- coloured species. As a result of a redox reaction, Cr (VI) is reduced to Cr (III), and DPC is oxidized to 1,5-diphenylcarbazone (DPCA). Chromium (III) and DPCA form a purple-coloured species with lambda max of 540 nm (Onchoke and Sasu, 2016).
Genomic DNA was extracted from efficient chromium resistance isolates according the method described by Tillett and Neilan (2000). To confirm the species of bacterial isolates at the molecular level, the 16S rDNA genes of the five isolates were amplified by PCR using the universal primers (reverse primer: 5’-GAGAGTTTGATCCTGGCTGGCTCAG-3’ and forward primer: 5’-AAGGAGGTGATCCAGCCGCA-3’) according to Kang et al. (2010). The PCR was performed in the thermal cycle 2720 (Applied Biosystem, Waltham, MA, USA) in a total volume of 25 μL containing 1 μL of each primer, 3 μL of template DNA (50 ng/μL), 12.5 μL of 1× PCR master mix (GeneDireX) and 7.5 μL of water nuclease-free). Amplification of 16S rDNA gene was executed under the following conditions: initial denaturation at 94°C for 5 min followed by 35 cycles of 1 min denaturation at 94°C, 1 min primer annealing at 52°C, 1 min extension at 72°C and final extension at 72°C for 10 min.
The PCR products were purified using Montage PCR Clean up Kit (Millipore) following manufacture instructions to remove unincorporated PCR primers. The PCR products of approximately 1,400 bp were subjected to sequencing through lab technology services located in Korea and performed at Applied Biosystems model 3730XL automated DNA sequencing system. The GenBank Accession numbers for the 16S rDNA gene sequence of Pseudomonas aerginosa (NCBI code: FAYP2) and P. zhadogens (FAYP3) isolates were OM010239 and OM019074 respective.
Extraction of the plasmid DNA from all isolates was carried out using alkaline lysis method (Shakibaie et al., 2009). Curing experiments were performed using 10% SDS and elevating temperature at 45°C as curing agents. Briefly, overnight culture of Pseudomonas isolates was grown in presence of sub- inhibitory concentration of curing agents for 24 h at 37°C. A loop full of the organism from the highest concentration of curing agents streaked on LB agar to obtain isolated colonies. In case of high temperature, the overnight growth of the organism was streaked on LB agar plates and incubated at 45°C for 48 h. The individual colonies were inoculated and incubate at the same temperatures. The physical loss of plasmid in the cured derivatives was confirmed by agarose gel electrophoresis (Lee et al., 1994).
RESULTS
A total of 50 bacterial isolates were obtained from different leather tanneries area which collected from various locations in Fayoum governorate, Egypt. The isolates were maintained on nutrient agar plates till further studies. These isolates were screened for their morphological properties to obtain the more closely related Pseudomonas isolates, Only 12 isolates out of 100 isolates showed positive test for morphological and biochemical tests.
The twelve isolates FAYP1, FAYP 2, FAYP 3, FAYP 4, FAYP 10, FAYP 11, FAYP 13, FAYP 15, FAYP 18, FAYP 20, FAYP 23, and FAYP 28 were more resistant than the others grown on nutrient broth containing 100 μg Cr (VI)/mL. The isolates No. FAYP2, FAYP3, FAYP18, FAYP20, and FAYP13 observed the highest significant growth compared with other isolates, which indicates that this isolate is tolerant to the Cr (VI) at concentration (100 μg/mL). The results in Table 1 showed the reduction percent of chromium Cr (VI) (100 μg/mL) for the isolates. Based on previous assessments, two isolates FAYP2 and FAYP3 were selected for further studies as hexavalent resistant bacterial isolates.
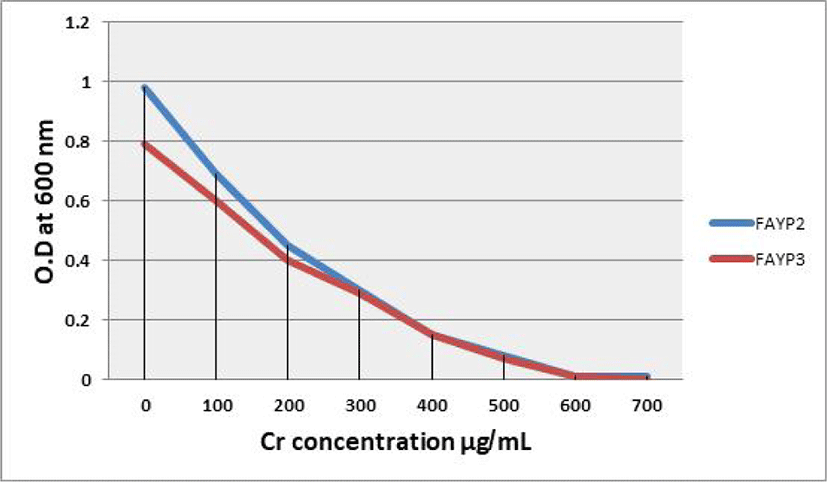
The effect of different Cr (VI) concentrations on the growth of the two selected isolates FAYP1 and FAYP2 in liquid medium was variable. Figs. 1 and 2 were showed the relationship between the growth of cells and initial Cr (VI) concentrations. The bacterial growth was decreased consistently with increasing concentrations of Cr (VI). The most significant growth decreased after addition of 500 μg/mL of Cr (VI) was observed with isolate FAYP2. However, isolate FAYP3 was affected by the chromium concentration added (600 μg/mL). Isolates FAYP2 and FAYP3 grow well when exposed to the highest chromium concentration, up to till 200 μg/mL. The detoxification efficiency of the two respective isolates follows the sequence: FAYP2>FAYP3. The two isolates (FAYP2 and FAYP3) were selected for further experiments.
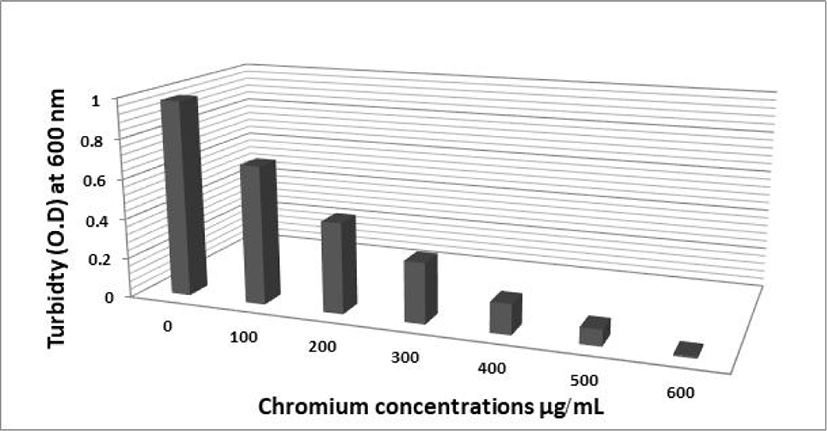
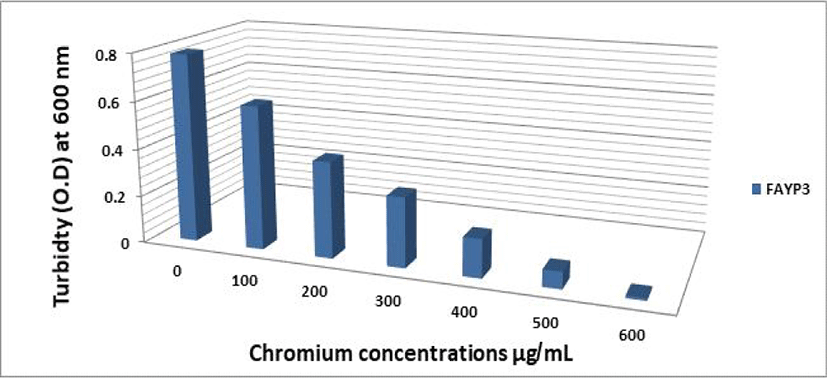
| No | Isolates code | Cr VI reduction (%) |
|---|---|---|
| 1 | FAYP2 | 70 |
| 2 | FAYP3 | 60 |
| 3 | FAYP18 | 55 |
| 4 | FAYP20 | 50 |
| 5 | AYP1 | 45 |
| 6 | AYP23 | 40 |
| 7 | AYP4 | 37 |
| 8 | AYP15 | 34 |
| 9 | AYP28 | 32 |
| 10 | AYP10 | 29 |
| 11 | AYP5 | 26 |
| 12 | AYP13 | 24 |
Fig. 3 shows the result of MIC of chromium (IV) by the two selected isolates FAYP2 and FAYP3. The growth of bacteria declined with increasing concentration of chromium. The two bacteria tested showing no growth at nutrient agar medium contain 600 μg/mL Cr (IV). The two isolates tested have a visible growth at 400 μg/mL because the Cr (IV) concentration still can be resisted by all bacteria. All bacteria tested show poor growth at medium containing 500 μg/mL Cr (IV) and showing the total inhibited growth at medium containing 600 μg/mL Cr (IV). Bacteria cannot grow at the 600 μg/mL concentration of Cr (IV)because the bacteria tested can only resist under 600 μg/mL Cr (IV).
Using Beer’s Law, the absorbance of 1,5-diphenylcarbazide (DPC) at different concentration of standard Cr (VI) was plotted to give a linear curve. Linear regression line or least square method was applied to obtain the best straight line (Fig. 4) through the new slope and intercept as shown from the equation, Y = 1.4053 × 104 X + 1.425 × 10−3. As a result of a redox reaction, Cr (IV) is reduced to Cr III and DPC is oxidized to 1,5-diphenylcarbazone (DPCA and formed a purple-color. The formed purple color complex indicates the presence of Cr (VI) which is measured spectrophotometry at 540 nm. The results in Table 2 showed the positive relationship between the concentration of hexavalent chromium and the absorbance value of DPC, when the hexavalent chromium concentration decreased the absorbance of DPC value will be decreased. The growth of bacterial isolates was correlated with their capacities to reduce Cr (IV). Isolates FAYP2 and FAYP3 gave the highest growth and the highest Cr (IV) reduction. The chromium (VI) reduction efficiency by bacterial isolates decreased by increasing the Cr (VI) concentration in the medium. The correlation between the bacterial growth and Cr (IV) illustrate the percentage of Cr (IV) reduction to Cr (III) by different bacterial isolates is presented in Fig3. The influence of initial Cr (VI) concentrations on the percentage of Cr VI reduction by FAYP2 and FAYP3 isolates were showed in Table 3.

| Chromium (VI) concentration (μg/mL) | Absorbance of DPC | |
|---|---|---|
| Isolate FAYP2 | Isolate FAYP3 | |
| 100 | 0.0251 | 0.0291 |
| 200 | 0.0413 | 0.0583 |
| 300 | 0.0765 | 0.0951 |
| 400 | 0.1375 | 0.1517 |
| 500 | 0.2021 | 0.2143 |
| Initial chromium concentration (μg/mL) | Final chromium concentration (μg/mL) | Chromium removal (%) | |
|---|---|---|---|
| FAYP2 isolate | FAYP3 isolate | ||
| 100 | 30 | 70 | 60 |
| 200 | 70 | 65 | 52 |
| 300 | 129 | 57 | 47 |
| 400 | 220 | 45 | 40 |
| 500 | 325 | 35 | 31 |
The few morphological characters with limited variations may lead to an overlap and misidentification of the strains, so the morphological and biochemical identification of the isolates were confirmed by molecular identification based on 16S rDNA gene analyses. PCR amplification of 16S rDNA gene of the two isolates of Pseudomonas produced a single fragment of 1,400 bp by gel electrophoresis that determined compared with the related sequences in Genbank. The representative 16S rDNA gene sequences of Pseudomonas strains were chosen for phylogenetic and comparison with other strains. Each sequence after editing was submitted to the GenBank and homology searches were done of all Pseudomonas sequence basic local alignment search tool (BLAST) nucleotide and FASTA programs (National Center for Biotechnology Information (NCBI), Bethesda, MD, USA). BLAST search results of each sequence giving the closest match to the test sample was used to determine the species of Pseudomonas strains.
The 16S rRNA molecular were published on the NCBI website where BLAST tools and the phylogenetic analysis. Figs. 4 and 5 revealed that bacterial isolates FAYP2 and FAYP3 were identified as P. aeruginosa and P. zhadogens respectively. The phylogenetic tree obtained by sequence analysis of 16S rDNA of the two isolates of Pseudomonas strains and the other sequence of Pseudomonas strains that obtained from sequence data banks.

A plasmid of approximately 2 kb in size was detected in the two Pseudomonas strains. The chromium reduction present was measured before and after plasmid curing and it found 70% and 13.5% respectively, and the reduction percentage was decreased in the curing strains compared with wild type strain (non-cured) and the results indicate that the genes responsible for reduction most located on plasmid DNA.
Discussion
Bioremediation is the process of ecologically degrading organic wastes under conditions to benign state or levels below regulatory concentration limits. Because microorganisms have enzymes that allow them to reduce the environmental toxins, they are well-suited to the task of contaminant destruction (Kumar et al., 2011). Bioremediation has the advantage of being a natural process that takes a short amount of time and is an acceptable waste treatment procedure for polluted materials such as soil. Microbes that can decompose the contamination and multiply as a result of its presence (Verma and Jaiswal, 2016). The bio derivative population decreases as the pollutant is degraded loses less than other conventional ways for hazardous waste cleanup. The effectiveness of bioremediation depends on many factors; including, the chemical nature and concentration of pollutants, the physicochemical characteristics of the environment, and their accessibility to existing microorganisms It also aids in the full elimination of pollutants; many dangerous substances can reconverted into harmless products, and this aspect reduces the possibility of future liability related with the treatment compounds (El Fantroussi and Agathos, 2005). Heavy metals are removed from aqueous solution by bacteria, fungi, ciliates, algae, mosses, macrophysics, and higher plants by a variety of methods rapping into cellular capsules, precipitation and oxidation-reduction events, as well as the production of protein-DNA adducts are all part of the biological response to metals (Montagnolli et al., 2015).
Chromium (VI) is one of the major pollutants released from tannery industry; dyes and textile industry waste effluents and were highly toxic and carcinogenic in nature. Hexavalent chromium Cr (VI) is associated with various forms of cancer particularly pancreatic cancers (Alguacil et al., 2004) and respiratory tract (Kuo et al., 2006). In the present investigation, twelve chromium tolerant isolates were isolated and their chromium tolerance limits was determined. Out of these five isolates have been found potential tolerant to elevated chromium concentration of up to 600 μg/mL. The chromium tolerant microorganisms were identified as Pseudomonas in accordance with the Berge’s manual of determinative bacteriology.
During the present investigation two isolates were selected FAYP2 and FAYP3 according to their highly resistant to chromium at a concentration of 500 and 600 μg/mL, respectively. Chromium (VI) is a common pollutant introduced into natural waters from a variety of industrial effluents and its removal by reduction has been well documented (Pattanapipitpaisal et al., 2002; Sultan and Hasnain, 2003). In the present study both selected isolates FAYP2 and FAYP3 could reduce Cr (VI) (100 μg/mL) 70% and 60% from the medium after 96 h, respectively. One potential method is microbial catalyzed reduction of Cr (VI) to Cr (III), which was first reported with Pseudomonas spp. (Romanenko and Koren’Kov, 1977). Since then, significant progress has been made towards understanding the processes controlling enzymatic reduction of Cr (VI) in Gram-negative bacteria, especially those belonging to the genus Pseudomonas (Chardin et al., 2003).
Studies developed by Park et al. (2005) suggested that Cr (VI) can be reduced to Cr (III) by the biomass through two different mechanisms: in the first mechanism, Cr (VI) is directly reduced to Cr (III) in the aqueous phase by contact with the electron-donor groups of the biomass, and the second mechanism consists of three steps: (1) binding of anionic Cr (VI) ion species to the positively charged groups present on the biomass surface; (2) reduction of Cr (VI) to Cr (III) by adjacent electron-donor groups; and (3) release of the Cr (III) ions into the aqueous phase due to electronic repulsion between the positively charged groups on the cells surface and the Cr (III) ions, or the complexion of the Cr (III) with adjacent groups capable of Cr-binding. The ability to reduce Cr (VI) and to resist high Cr (VI) concentrations were found to be independent properties of bacteria especially Pseudomonas sp. The Cr (VI) reduction of the isolates was expressed as Cr (VI) and Cr (III) concentrations in the medium. P. aeruginosa and P. stutzeri isolates were exposed to 10 mg/L chromium for 48 h in AMM medium (Christl et al., 2012).
The bacterial isolates were then characterized by morphological, biochemical tests, multiple heavy metal resistance capacity, MIC and comparative heavy metal degradation capacity. Depending on gram staining, two isolates (FAYP2 and FAYP3) were identified as gram-negative based on their morphological and biochemical characterization. These isolates were further subjected for the phylogenetic analysis by 16S rRNA gene analysis. Thus based on the 16S rRNA analysis the two strains were identified as follows: FAYP2- Pseudomonas areuginosa and FAYP3- P.zhadogens However, when the sequences were subjected to BLAST search, this FAYP2 and FSYP3 shows a sequence homology of 99% with P. areuginosa and P.zhadogens, respectively.
Relative effects of bacterial growth in presence of chromium in different concentrations (100–1,200 μg/mL) were studied and it was observed that bacterial growth is concentration dependent, since it showed decreasing optical density in accordance with the increasing chromium concentration. MIC is the lowest concentration at which the isolate is completely suppressed (as demonstrated by the absence of visible bacterial growth) is recorded. In this study the MICs for the isolates FAYP2 and FAYP3 was found to 600 μg/mL. Khare et al. (1997) have reported that due to stress, 50% of the bacterial population of the tannery sediment consisted of Pseudomonas, indicating that the stress factors present in the tannery waste had reduced the species diversity considerably. The Pseudomonas populations which were present in the tannery sediment tolerated a concentration up to 50 mg Cr VI/L. At a higher concentration of 150 mg Cr VI/L, 80% of the Pseudomonas population was inhibited (Khare et al., 1997).
Bacterial plasmids encode resistance systems for toxic metal ions are inherited by plasmids in several species of bacteria especially Pseudomonas syringae and Escherichia coli and documented by several authors (Brown et al., 1995; Silver et al., 1993; Williams et al., 1993). Singh et al. (2010) showed heavy metal resistance capacity either plasmid mediated or chromosomal DNA mediated. For determination of genetic basis for metal resistance, plasmid profiling is important. Plasmid DNA extraction of two bacterial isolates having biodegradation capacity was assessed to understand whether their chromium resistance capacity is plasmid DNA or chromosomal DNA mediated. In our study, the selected isolates FAYP2 and FAYP3 showed plasmid DNA. In bacteria, the heavy metal resistant genes are located either on the bacterial chromosome or in the plasmids or on both (Nies and Brown, 1998). According to Malik (2004), Cd and Cr resistant genes are present in plasmid DNA but Pb resistance gene is located on chromosomal DN A of Enterobacteria.
In many studies, loss of plasmid completely disrupted tolerance to heavy metals. The pseudomonas spp. investigated in the current study also harbors a plasmid and the curing of this plasmid was caused partially inhibition of chromium tolerance. Rather chromium tolerance efficiency of the plasmid cured cells was decreased as compared to the wild-type cells. This suggests that in this Pseudomonas genes associated with chromium tolerance are distributed on both chromosomes and plasmids (Edward Raja and Selvam, 2009).
Conclusion
In conclusion, the heavy metal degrading potentiality of samples taken from tannery industrial settings in three polluted locations was examined in this study. Two bacterial isolates were isolated from those samples; the isolates were tested for chromium Cr IV resistance capability and growth curve analysis. The isolates were identified as Pseudomonas spp. based on several biochemical characterization assays. Our results support the idea that the bacterial isolates FAYP2 and FAYP3 have high bioremediation potential and might be exploited to develop bioremediation agents to detoxify tannery effluents in industrial waste waters.
